As anyone following the field will know, the derivation of induced pluripotent (iPS) cells reprogrammed from differentiated somatic cells offers a remarkable promise: the ability to generate donor-specific pluripotent stem cells, without the “ethical” confusion that has so unfortunately retarded the progress of somatic cell nuclear transfer (SCNT) research. However, protocols to date have not led to systems that could yet viably be scaled up into therapeutic use: efficiency has been extremely low, and the reliance on the proto-oncogene c-Myc and/or of viral vectors in such systems have made the risk of transformation too high to realistically be tested in humans. A more detailed understanding of the mechanistic basis for reprogramming could potentially allow for the bypassing of these steps, and/or increases in efficiency, that would bring rejuvenation therapies (and thus treatments for the degenerative processes of aging and a range of injuries and diseases) closer to the clinic.
We were therefore surprised to note how little attention has been given to a recent report in Nature(1) from the Blau lab at Stanford’s Institute for Stem Cell Biology and Regenerative Medicine. In the few commentaries that have appeared online, either offhanded allusion or no discussion at all is given to the potential that these findings could lead to tools that would greatly increase the efficiency of cellular reprogramming, although a recent review(7) was more appropriately enthusiastic in reviewing this and a related report (9).
Their group and others had previously shown that fusing mouse ESC with human fibroblasts led to heterokaryons led to induction of previously-silent genes that allowed the cells, like iPS cells, to take on other differentiated cell fates. They “showed that reprogramming in heterokaryons was influenced by DNA methylation status, tissue of origin, and the relative ratio of nuclei that dictates the balance of regulators, consistent with recent experiments in iPS cells.” (1) In the present study, they exploited this system
to study epigenetic and transcriptional changes critical to the initiation of reprogramming towards pluripotency. We focused on DNA demethylation—a known block to reprogramming that leads to partially reprogrammed iPS cells, and also a key step for reprogramming by nuclear transfer. Despite decades of effort, so far no consensus mammalian DNA demethylase has been identified. Recently, [Activation-Induced (Cytidine) Deaminase (AID)] has been implicated in DNA demethylation in zebrafish within hours after fertilization, acting in a complex that mediates deamination followed by DNA repair. In mammals, AID is primarily known for its role in the generation of antibody diversity in B lymphocytes, but has recently been detected in germ cells. (1)
Indeed, in addition to germ cells,(2,3) AID had been found to be activated in oogenesis and early development,(4) including during spermatogenesis in both normal and telomerase-disabled (TERC-/-) mice.(5)† It was therefore a reasonable candidate for involvement in maintenance or induction of pluripotency. The hetereokaryons seemed likely to be a good system in which to study such changes during the presumed reprogramming process because interspecies differences would allow them to distinguish between expression changes in gene transcripts derived from the two parent cell types; moreover, the process does not involve cell division, allowing them to rule out the possibility that any observed CpG demethylation might be the result of stochastic errors in methylation maintenance after replication of DNA during S phase.
(siRNA)… showed that… AID … is required for promoter demethylation and induction of OCT4 (also known as POU5F1) and NANOG gene expression. AID protein bound silent methylated OCT4 and NANOG promoters in fibroblasts [and the homeobox protein Cdx2 in mouse ESC], but not active demethylated promoters in ES cells. These data provide new evidence that mammalian AID is required for active DNA demethylation and initiation of nuclear reprogramming towards pluripotency in human somatic cells.(1)
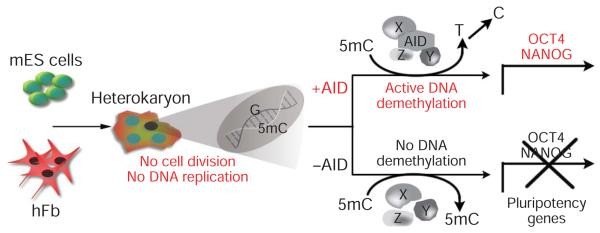
Figure: “Model for AID-dependent active DNA demethylation in reprogramming. … The other putative components of this mammalian DNA demethylase complex (X, Y and Z) that may act together with the deaminase, AID, remain to be identified. 5mC, 5-methyl-cytosine.” From (1).
As was noted by a commentary by Drs. Suneet Agarwal and George Q Daley,
Several important questions remain: (1) Are the same mechanisms at work during reprogramming by NT [somatic cell nuclear transfer] or in the generation of iPS cells? The possibility is favored by the observations of active demethylation of the paternal genome after natural fertilization, and the facilitation of iPS cell generation using agents that interfere with maintenance of DNA methylation. … The results of Bhutani et al. suggest that AID function is not replaceable by other cytidine deaminase family members. Therefore, it would be interesting … to assess the effects of AID disruption on human iPS cell generation … (2) What are the mechanisms activating AID during reprogramming? The observation by Bhutani et al. that AID binds to the OCT4 and NANOG promoters in fibroblasts, where the loci remain methylated, implies that the presence of AID is not sufficient for demethylation. Factors either resident in mouse ES cells or induced in the heterokaryons must therefore be invoked to explain AID-mediated activation of demethylation at these loci after fusion.(8)
Again, we are surprised by the mild tone and narrowly-defined discussion of the potential of this report. Perhaps the remarkable rush of research monies and investigators into the iPS field, and the ensuing breathtaking pace of progress in iPS research that began in 2008 and has continued ever since, has led to some unfortunate fatigue with the sheer volume of reports. Indeed, the very same issue of Nature also featured a supporting report that “Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency,”(9) and the potentially-useful finding that “Tbx3 improves the germ-line competency of induced pluripotent stem cells”(10) (which seems to have garnered more enthusiasm for its clinical potential). Whatever the reason for the blasé response, small molecules or other convenient and readily-reversible inducers of the same processes could reasonably be anticipated to lead to a similar leap in efficiency and rapidity of reprogramming similar to that observed in the Blau group’s heterokaryons, bringing the safe and scalable production of patient-specific pluripotent cells needed for cell therapy in tissue engineering for biomedical rejuvenation closer to reality.
†The expression of AID during spermatogenesis in telomerase-deficient mice,(5) combined with its involvement in somatic hypermutation and immunoglobulin class switching in B-cells, and the observed elongation of B-cell telomeres in a heterogeneous pattern in the same model following immunization(6) and several other findings, led Dr. de Grey to propose that aberrant AID expression might be a key component of the ALT telomere-lengthening mechanism. SENS Foundation funded preliminary studies to probe this possibility by Dr. F. Mathias Bollmann at Universitätsklinikum Hamburg-Eppendorf; these studies appear to have discounted AID’s involvement (unpublished results; personal communication, M. Bollmann).
References
1. Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010 Feb 25;463(7284):1042-7. PubMed PMID: 20027182.
2. Schreck S, Buettner M, Kremmer E, Bogdan M, Herbst H, Niedobitek G. Activation-induced cytidine deaminase (AID) is expressed in normal spermatogenesis but only infrequently in testicular germ cell tumours. J Pathol. 2006 Sep;210(1):26-31. PubMed PMID: 16783758.
3. Morgan HD, Dean W, Coker HA, Reik W, Petersen-Mahrt SK. Activation-induced cytidine deaminase deaminates 5-methylcytosine in DNA and is expressed in pluripotent tissues: implications for epigenetic reprogramming. J Biol Chem. 2004 Dec 10;279(50):52353-60. Epub 2004 Sep 24. PubMed PMID: 15448152.
4. Liu L, Bailey SM, Okuka M, Muñoz P, Li C, Zhou L, Wu C, Czerwiec E, Sandler L, Seyfang A, Blasco MA, Keefe DL. Telomere lengthening early in development. Nat Cell Biol. 2007 Dec;9(12):1436-41. Epub 2007 Nov 4. PubMed PMID: 17982445.
5. Tanemura K, Ogura A, Cheong C, Gotoh H, Matsumoto K, Sato E, Hayashi Y, Lee HW, Kondo T. Dynamic rearrangement of telomeres during spermatogenesis in mice. Dev Biol. 2005 May 15;281(2):196-207. PubMed PMID: 15893973.
6. Herrera E, Martínez-A C, Blasco MA. Impaired germinal center reaction in mice with short telomeres. EMBO J. 2000 Feb 1;19(3):472-81. PubMed PMID: 10654945; PubMed Central PMCID: PMC305584.
7. Deng W. AID in reprogramming: quick and efficient: identification of a key enzyme called AID, and its activity in DNA demethylation, may help to overcome a pivotal epigenetic barrier in reprogramming somatic cells toward pluripotency. Bioessays. 2010 May;32(5):385-7. PubMed PMID: 20394066.
8. Agarwal S, Daley GQ. AID for reprogramming. Cell Res. 2010 Mar;20(3):253-5. PubMed PMID: 20190775.
9. Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010 Feb 25;463(7284):1101-5. PubMed PMID: 20098412.
10. Han J, Yuan P, Yang H, Zhang J, Soh BS, Li P, Lim SL, Cao S, Tay J, Orlov YL, Lufkin T, Ng HH, Tam WL, Lim B. Tbx3 improves the germ-line competency of induced pluripotent stem cells. Nature. 2010 Feb 25;463(7284):1096-100. Epub 2010 Feb 7. PubMed PMID: 20139965.



