My name is Summer Wang, and I am a rising senior at the University of Washington, where I am majoring in Molecular, Cellular, and Developmental Biology and minoring in Chemistry. Under the mentorship of Bonita Brewer and M.K. Raghuraman at the University of Washington, I am attempting to identify cellular pathways and mechanisms that can regulate ribosomal DNA (rDNA) copy number in the budding yeast Saccharomyces cerevisiae. The rDNA region, which encodes ribosomal RNA that is a key component of the millions of ribosomes in the cell, contains 150 tandem repeats in S. cerevisiae [1]. Previous work demonstrated correlations between altered rDNA sizes and genome-wide DNA replication as well as replicative lifespan [2,3]. We proposed a model whereby rDNA copy number is balanced between genome replication and ribosomal demands. We hypothesize that too few rDNA copies cannot satisfy ribosome demand while too many copies may interfere with genome-wide DNA replication and that certain genetic pathways must function to maintain such balance.
To identify genetic regulators of rDNA copy number, I screened 400 mutants for rDNA size. Two hundred of the strains tested are documented to have longer replicative lifespan while the remainders of the test panel were randomly selected from a yeast deletion collection (YKO collection) [4]. Statistically, mutants with longer lifespan do not have different rDNA size than the random mutants that are not long-lived. I still want to look for genetic regulators of rDNA size or common pathways that can influence rDNA flexibility. Therefore, 64 mutants with rDNA sizes that significantly differ from the mean by at least one standard deviation were picked for further study. By analyzing the functions of these 64 mutants and the cellular pathways that they are involved in, I found three intriguing common biological functions.
When I return to the University of Washington in the fall, I will continue creating de novo knockout strains to rule out the possibility that the altered rDNA copy number is caused by random mutations that took place in the maintenance of the collection. Once we have confirmed a link between a specific pathway and rDNA copy number, I can test more mutants that share similar biological functions or share a common biological pathway to see if they influence rDNA copy number. I am also investigating how ploidy affects rDNA copy number maintenance since it is poorly understood. We hypothesized that the rDNA copy number may change over divisions. However, my preliminary data suggests that diploid cells can maintain longer or shorter rDNA sizes relatively stably. I believe that studying the regulation of rDNA loci not only can lead me to the genetic regulators of rDNA size but also can provide new insight into such cellular mechanisms as DNA replication.
Rescue of mitochondrial dysfunction by transferring mitochondrial DNA-encoded genes to the nucleus
This summer, I will be working in the mitochondria group in SENS Research Foundation. One of the MitoSENS group’s goals is to use engineered mitochondrial genes to rescue cells with compromised mitochondrial function. Mitochondria serve as the energy source in all eukaryotic organisms. This organelle contains 1000 – 1500 proteins, and the majority of them are encoded in the nucleus and transported into mitochondria. Mitochondrial DNA (mtDNA) is around 16 kb in length and encodes 13 different proteins [5]. Since mitochondria do not possess a substantial DNA repair system, mitochondrial DNA mutations accumulate over time and can lead to respiratory chain defects, which can lead to cell survivorship problems due to insufficient energy production.
Currently, researchers are trying to develop gene therapies in order to treat human diseases caused by mitochondrial DNA mutations, but available methods are limited. One of the approaches involves inserting the wild-type copy of a gene normally found in the mitochondria into the nucleus. The missing mitochondrial function can then be rescued by targeting the protein to mitochondria. Specifically, the MitoSENS research group’s ultimate goal is to insert the 13 mitochondrial DNA-encoded genes into the nuclear genome, target the proteins to the mitochondria, and replace the loss of function of those genes. If the mRNA is exported from the nucleus into the cytoplasm where it is translated while attached to the mitochondria outer membrane, then the protein can be transported into the mitochondria and serve its normal biological function.
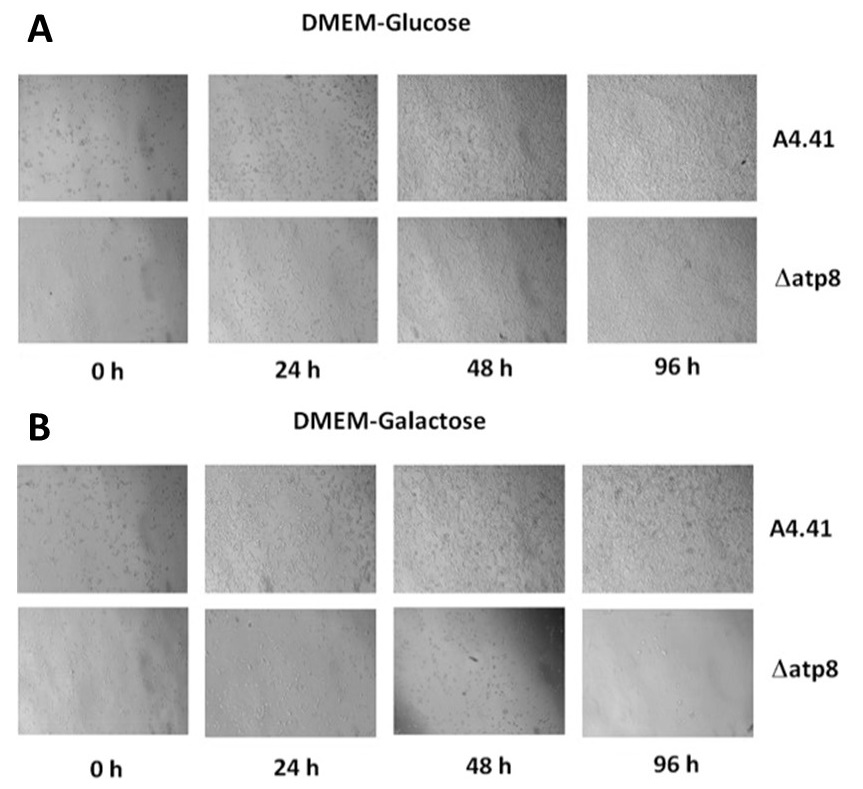
Figure 1. ATP8 is required for mitochondrial oxidative phosphorylation.
A. Both normal cells (A4.41) and ATP8-null cell lines (Δatp8) can grow in glucose medium as evidenced by the cell monolayer.
B. Cells need to use mitochondrial oxidative phosphorylation to provide ATP in galactose medium. Normal cells can grow while Atp8-null cell lines die after 4 days as evidenced by the lack of cells in visible in the final panel.
My project is focusing on two proteins that are encoded by the mtDNA: ATP6 and ATP8, which are subunits in complex V in the respiratory chain. Previous work from the Mafredi group on ATP6 showed rescue of ATP synthesis deficiency [6]. Using a construct similar to the one used by Mafredi et al., preliminary data from our group showed stable expression of ATP8, another mitochondrial DNA-encoded gene, in ATP8-null cells. We want to improve upon this work by developing a more effective construct with optimized protein expression and then fully rescue the missing mitochondrial function (Figure1). For example, Ali Crampton, a previous intern, determined that the untranslated sequence from ATP5G2, another gene involved in mitochondrial function, provided the most promising mitochondrial targeting to-date. By using the mitochondria target sequence region of this gene, we are hoping to improve import of ATP8 protein into ATP8-null cells, which may lead to normal complex V structure and ultimately rescue mitochondrial function.
Future Plans:
My plan is to attend graduate school after college. I want to pursue a PhD degree in the field of molecular cell biology, so I can keep researching in this limitless kingdom of biology. My research interests have always centered on cellular mechanisms and genetics, such as DNA replication and cell signaling. I believe that the key to understanding most mysteries in nature lies in studying the most basic molecular pathways.
References:
1. Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12:3821–3830.
2. Kwan, E. X., Foss, E. J., Tsuchiyama, S., Alvino, G. M., Kruglyak, L., Kaeberlein, M., Raghuraman, M. K., Brewer, B. J., Kennedy, B. K., and Bedalov, A. A Natural Polymorphism in rDNA Replication Origins Links Origin Activation with Calorie Restriction and Lifespan. PLoS Genet 2013.
3. Ide S, Miyazaki T, Maki H, Kobayashi T. Abundance of ribosomal RNA gene copies maintains genome integrity. Science. 2010 Feb 5; 327(5966): 693-6.
4. McCormick MA, Kennedy BK. Genome-scale studies of aging: challenges and opportunities. Curr Genomics. 2012 Nov; 13(7): 500-7.
5. Cwerman-Thibault H1, Sahel JA, Corral-Debrinski M. Mitochondrial medicine: to a new era of gene therapy for mitochondrial DNA mutations. J Inherit Metab Dis. 2011 Apr;34(2):327-44.
6. Manfredi G1, Fu J, Ojaimi J, Sadlock JE, Kwong JQ, Guy J, Schon EA. Rescue of a deficiency in ATP synthesis by transfer of MTATP6, a mitochondrial DNA-encoded gene, to the nucleus. Nat Genet. 2002 Apr;30(4):394-9.


