As we reviewed in a previous posting on a recent advance in genetic engineering with zinc finger nucleases (ZFNs),
In addition to its widely-anticipated potential to provide highly-effective therapies for genetic disorders, somatic gene therapy is an essential enabling technology for the repair or obviation of several of the cellular and molecular lesions driving age-related disease and dysfunction (notably the accumulations of mutations in mitochondrial and nuclear DNA [including the medium-term obviation of the latter through WILT]). One of the most promising routes to somatic gene therapy is zinc finger nucleases (ZFNs), engineered DNA-binding proteins consisting of a FokI restriction enzyme catalytic core bookmarked into a dimer of zinc finger array DNA binding domains. The choice of zinc finger domains allows the engineer to target twinned 9-18 base-pair sequences in the recipient genome, separated from each other by a (typically) 5-7 base pair spacer. Upon binding, the restriction enzyme dimerizes, creating a double-strand break at the spacer locus; the engineer then takes advantage of the native DNA repair machinery to insert an user-supplied DNA repair template through Non-Homologous End Joining (NHEJ).
Despite the many advantages of ZFNs, and the landmark 2009 report of their use to generate precision knockout mice,(1) relatively little use has been made of ZFNs for either basic science or translational work toward gene therapies. This is principally because of the difficulty and expense of generating novel ZFN binding arrays, whose creation using the standard technique of modular assembly is often unsuccessful due to combinatorial incompatibilities amongst individual “finger” peptides which, in sequence, would otherwise be expected to bind to the targeted DNA sequence: when placed in sequence, such peptides are often found to interfere with one another’s DNA-binding capacity.
To overcome these difficulties without recourse to expensive and (some argue) overly-restrictive purchase from patent-holder Sangamo BioSciences (via its licensee, Sigma-Aldrich), a group of researchers centered at Massachusetts General Hospital and the University of Minnesota united to form the Zinc Finger Consortium, to create a new platform for ZFN creation on “open source” principles, “committed to developing resources, software, and other tools for engineering zinc fingers and for performing genome engineering that are robust, user-friendly, and publicly available to the academic scientific community.” The first fruits of this collaboration, reported in 2008,(2) was OPEN (Oligomerized Pool ENgineering), “a rapid, publicly available strategy for constructing multifinger arrays.” Despite its claimed efficiencies, however, it remained labor-intensive and expensive to establish OPEN methods in a given lab and to generate individual ZFNs, requiring as it did that each site establish its own library of peptide variants and then screen the collection for suitable ‘fingers’ to reach into each desired target site.
Now, the Massachusetts-Minnesota collaboration has reported that they have developed “context-dependent assembly (CoDA), a platform for engineering ZFNs using only standard cloning techniques or custom DNA synthesis”:
With the CoDA approach, three-finger arrays are assembled using N- and C-terminal fingers that have been previously identified in other arrays containing a common middle finger (F2 units) (Fig. 1). CoDA can be implemented by using a large archive consisting of 319 N-terminal-end fingers (F1 units) and 344 C-terminal-end fingers (F3 units) engineered to function well when positioned adjacent to one of 18 fixed F2 units. Thus, in contrast to modular assembly, CoDA does not treat fingers as independent modules but instead explicitly accounts for context-dependent effects between adjacent fingers, thereby increasing the probability that a multifinger array will function well. CoDA is rapid and requires neither specialized expertise nor labor-intensive selections; dozens of multifinger arrays can be constructed in 1–2 weeks or less using standard cloning techniques or commercial DNA synthesis.(3)
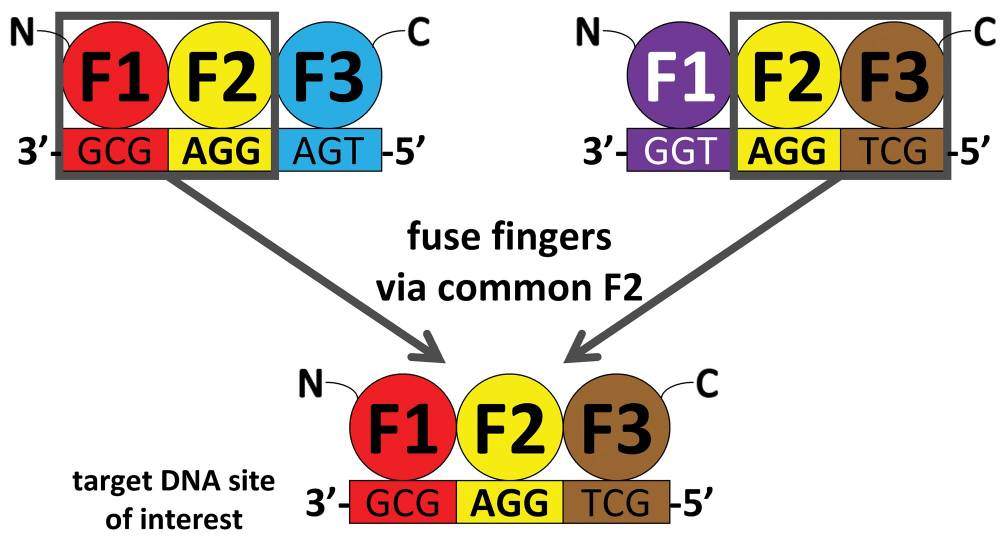
Figure 1. Schematic Overview of CoDA Method for Engineering Zinc Finger Nucleases. From (3).
If this is indeed the experience at independent labs — that they can use CoDA in an essentially off-the-shelf way, to rapidly identify and generate novel ZFNs in-house — then CoDA promises to greatly increase the power of genetic engineering, putting the unprecedented precision of ZFNs in genetic engineering of mammalian systems into the hands of basic researchers and opening up the potential of new gene therapies for recognized congenital diseases, while establishing an enabling technology that will be essential to future rejuvenation biotechnologies. It will also be a powerful validation of the optimism that many academic scientists and DIYbiohackers have poured into open-source biological and technological research and development. We may all some day owe these scientists a great debt, even if — especially if — we ultimately rarely recall their names or their report because the technology is so well-established as to be taken for granted, a part of the biomedical background as invisible to future generations that reap its benefits in health and longevity as sanitation systems and the absence of endemic polio are to the developed world today.
References
1. Geurts AM, Cost GJ, Freyvert Y, Zeitler B, Miller JC, Choi VM, Jenkins SS, Wood A, Cui X, Meng X, Vincent A, Lam S, Michalkiewicz M, Schilling R, Foeckler J, Kalloway S, Weiler H, Ménoret S, Anegon I, Davis GD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Jacob HJ, Buelow R. Knockout rats via embryo microinjection of zinc-finger nucleases. Science. 2009 Jul 24;325(5939):433. PubMed PMID: 19628861; PubMed Central PMCID: PMC2831805.
2. Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, Unger-Wallace E, Sander JD, Müller-Lerch F, Fu F, Pearlberg J, Göbel C, Dassie JP, Pruett-Miller SM, Porteus MH, Sgroi DC, Iafrate AJ, Dobbs D, McCray PB Jr, Cathomen T, Voytas DF, Joung JK. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008 Jul 25;31(2):294-301. PubMed PMID: 18657511; PubMed Central PMCID: PMC2535758.
3. Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods. 2011 Jan;8(1):67-9. Epub 2010 Dec 12. PubMed PMID: 21151135.



