My name is Tianhan Deng, and I am a rising junior at the University of California, Berkeley. I am pursuing a degree in Molecular and Cellular Biology with an emphasis on developmental genetics and a degree in clinical psychology. I have been particularly interested in the field of neuro-oncology and how genetic profiles of each patient can be used to develop personalized treatments.
Prior to joining the SRF Summer Scholars Program, I was heavily involved in a research project in the lab of Dr. Petritsch in UCSF, aimed at understanding the biology of low-grade gliomas. Glial cells are responsible for the support and nourishment for our neurons and can be further subdivided into astrocytes and oligodendrocytes, Gliomas, which are tumors in glial cells, constitute 80% of primary malignant brain tumors, and astrocytomas and oligodendrogliomas refer to cancer of two subtypes of glial cells, respectively. Because of their slower growth and distinct biology from all other brain tumors, these two types of gliomas are given a distinct classification called “low-grade gliomas”. Although low-grade gliomas often are not as aggressive as glioblastomas, standard therapies can cause them to rapidly recur, gain a much more aggressive phenotype and eventually turn into WHO (World Health Organization) grade IV secondary glioblastomas. The poor understanding of how low-grade gliomas become more malignant through differentiation defects makes treatment particularly difficult and unavailable for patients.
Therefore, my research aims to resolve the current therapeutic standstill questioning how the differentiation defects can lead to these cancers. In order to seek a better treatment to stop their malignant progression, we need to understand exactly what biological process is disrupted when these defects occur. I based my research on the current hypothesis that these low-grade gliomas originated from oligodendrocyte progenitor cells (OPCs), which can differentiate into both oligodendrocytes and astrocytes that give rise to the tumors. Our hypothesis is that a driver mutation is causing this particular cell population to be locked into an undifferentiated state or even to dedifferentiate back into a more primitive cell type by disrupting other gene expression that determines cell identity in the brain. By understanding the differentiation defects and their subsequent consequences, we can apply treatments to hopefully reverse the defects.
Differentiation defect are caused by gene mutations, and we eventually honed in on one gene called IDH1 (isocitrate dehydrogenase 1) gene. It is believed to be the culprit of many gliomas, with more than 80% of low-grade glioma patients carrying a homozygous R132H missense mutation of this gene. Given its prevalence in gliomas, we believe this mutation is the initiator in many low-grade gliomas. In order to compare the cells which have the mutant genes with the cells with normal genes or cells with too many copies of a normal gene (which can also result in cancer, we isolated OPCs from mice brains and cultured the cells in a petri dish. Then, we made cells to express this mutated version of IDH1, overexpress the normal version, or use an empty vector as control. We performed an in-vitro differentiation assay to push the OPCs into different cell lineages, then examined the differentiated cells using immunofluorescence microscopy. We found that the cells with the IDH1 mutation are unable to differentiate into proper oligodendrocytes. In contrast, they differentiate into something similar to reactive astrocytes, an abnormal form of astrocytes indicative of brain lesions. A portion of OPCs also show resistance to differentiation altogether and are locked into this stem-like state. Blockage to differentiation is a key hallmark to cancer cells, therefore I hypothesized that these OPCs have gained the potential to become tumor cells. In order to test this hypothesis, we analyzed tumor growth of the modified OPCs in the brains of mice that are susceptible to tumor formation and have special genes that allow us to track the cell fate of OPCs in the brain (NG2-dsredCreER-p53 KO/KO mice). Considering their degree of proliferation in cell culture, these tumors grow slowly. Yet, when we isolated IDH1 mutated tumors from these mice, surprisingly a small number of neural stem cells (NSC) were detected. The implication of this finding is still under investigation.
Currently, we are using RNA-seq, which is a technique to detect differential gene expressions in a cell population, to examine the biology of these neural stem cells as well as determine the gene expression differences between IDH1 mut and IDH1 wild-type (unmutated) OPCs. Using RNA-seq will allow us to determine how gene expression was altered by the IDH1 mutation in the mouse brain and may offer insight into the surprising existence of the neural stem cell population. For example, we are interested in checking if re-implantation can generate another tumor consisting entirely of IDH1 mut-NSCs in contrast to the original tumors comprised of entirely IDH1 mut-OPCs. These studies could help us elucidate the definitive cell of origin and the tumor-initiating process for low-grade gliomas.
Exploring potential preclinical therapeutics using a novel lung-to-brain metastases model
My project this summer aims to create the best model for lung-to-brain metastasis. I will be working under the supervision of Dr. Susana Moleirinho in the lab of Dr. Kahlid Shah at the Brigham and Women’s Hospital and Harvard Stem Cell Institute.
Secondary brain metastases are a devastating condition, bearing a dismal prognosis. A large number of brain metastases originate from lung carcinomas, specifically non-small cell lung adenocarcinomas. Due to its complicated biology and tendency to metastasize, it remains one of the deadliest tumors in the field and has posed a great challenge in finding a cure.
A promising step toward finding a cure has been the discovery of TRAIL (Tumor Necrosis Factor-Related Apoptosis Inducing Ligand). TRAIL produce anti-tumor effects by causing tumor cells to essentially “suicide,” and its specificity for tumor cells but not healthy cells makes it a great therapeutic approach. Recently, several efforts have shown that TRAIL can induce cancer cells to die in a Death receptor 4/5 (DR4/5)-dependent fashion. However, this model requires some cell lines to be sensitized to TRAIL therapy by blocking epidermal growth factor receptor (EGFR) to help form death-inducing signaling complex (DISC). Therefore, the lab has developed a fusion protein composed of a EGFR-nanobody that targets EGFR and TRAIL. This fusion protein is abbreviated Enb-Trail and it has shown significant effects in multiple cell lines.
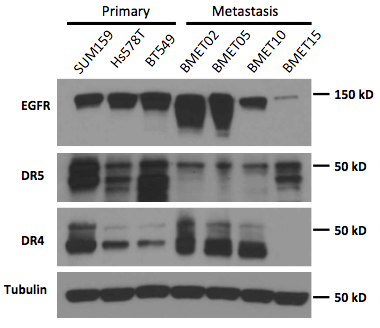
Figure 1. Expression of TRAIL-related Receptors.
Western blotting against several receptors related to our treatments: EGFR, DR4 and DR5 on several brain metastases and primary cell lines. We show that most of cancer cell lines express differential levels of DR4 and 5, validating the approach of using TRAIL as a therapeutic target. We also show the expressions of DR5 are generally downregulated in the secondary metastases, possibly as a mechanism for the cancer cells to escape TRAIL-induced apoptosis.
My project will focus on selecting a good model to recapitulate the biology of metastatic lung adenocarcinoma and gather the preclinical data for Enb-Trail on the brain metastases. To screen for the best model, we will treat several established lung adenocarcinoma cell lines, such as A549 and NCI-H23, with several targeted therapies to find the cell line(s) with sensitivity to our Enb-Trail therapy. We will perform several in-vitro cell-based assays to assess cell viability in response to treatments. Then, in order to verify their sensitivity or insensitivity to therapies, we will use a protein analysis protocol called a Western blot to further characterize the expression level of some key players in the targeted pathway, such as DR4, DR5, EGFR and CD36. Finally, in order to see how the therapy behaves in actual animals, we will put the model cells into the brains of mice through intracardiac injection to gather in-vivo efficacy data for the drugs in the brain, paying particular attention to its ability to cross the blood brain barrier (a barrier between peripheral blood and our cerebral-spinal fluid that prevents large molecules such as drugs from entering the brain).
In addition, we will also examine the potential of linking the TSR domain with TRAIL. The TSR domain is known for binding CD36 receptor and has been shown to be a potent inhibitor of angiogenesis (the process in which tumors build their own blood vessels). By targeting angiogenesis and apoptosis, the two hallmarks of cancer, at the same time, we may be able to identify a synergistic effect with the Enb-Trail treatment. If we observe a synergistic effect, we will explore the molecular mechanism between TSR and Trail to elucidate the interactions between these two proteins in their cooperation of fighting cancerous cells.
Future Plans:
Once the academic year resumes in the fall, I plan to continue my work in Dr. Petritsch’s lab. I also hope to return to Dr. Shah’s lab next summer to continue working on this Summer Scholar project. After graduation, I am planning on pursuing a MD/PhD dual degree program because the idea of translating basic research data into a clinical setting and being able to make a direct impact on patient care greatly interests me. I am hoping to continue pursuing neuro-oncology as my field of interest as well as becoming a neurosurgeon.


