Namaste! My name is Sumedh Anand Sontakke, and I am a junior majoring in electrical engineering at the College of Engineering, Pune, India. This summer, I will be working at the University of Oxford as a SRF Summer Scholar under the supervision of Professor Chas Bountra and Dr. David Brindley. I developed an interest in machine learning as a sophomore under the purview of computer vision, working on image processing and microcontroller-based robotics. Eventually, I came across bioinformatics – a field where data scientists and computer engineers like me could contribute to healthcare. It made me realize that I didn’t need a medical degree to make a difference in people’s lives.
I began my journey in bioinformatics at Skyline Labs, an FBStart-funded company. Here, I built a model to predict the occurrence of liver disease based on genetic and clinical trial data. The aim was to identify patients with a high risk of developing liver disease using machine learning algorithms. It may seem challenging to make such predictions without adequate medical qualification. But, imagine that the computer is a person with infinite patience who is able to learn from a number of experiences. A major difference between a physician and the computer is that the latter is able to learn from a large number of data points in a matter of hours: an impossible feat for the human mind. The computer makes all future decisions on the basis of these past experiences. The potential of the idea jumped out at me. In the forgotten corners of the world, where quality healthcare hasn’t percolated, such a tool could be useful in identifying high risk patients. This product would also significantly improve the accessibility of healthcare to people residing in the developed countries in the world. Instead of needing to travel great distances to consult experts, this tool could potentially allow any physician or sometimes even the patient to make a diagnosis.
The research was conducted using a type of classifier algorithm called an artificial neural network (ANN). An ANN’s mode of learning is similar to a primitive animal brain. It consists of ‘neurons’ which are the connections between the ‘layers’ of the model. When the computer is fed data, these neurons are developed akin to the way they are in an animal brain. The figure below illustrates the algorithm (Figure 1). In this example, the input layer is responsible for data collection and the two hidden layers are responsible for processing. The hidden layers are built by the computer and are where the conversion of input data to output data takes place. These and the output and input layers are all connected together by neurons, which are essentially just mathematical functions. The output of an ANN is a class label. Thus, in this case it was binary, indicating if a liver is healthy or not.
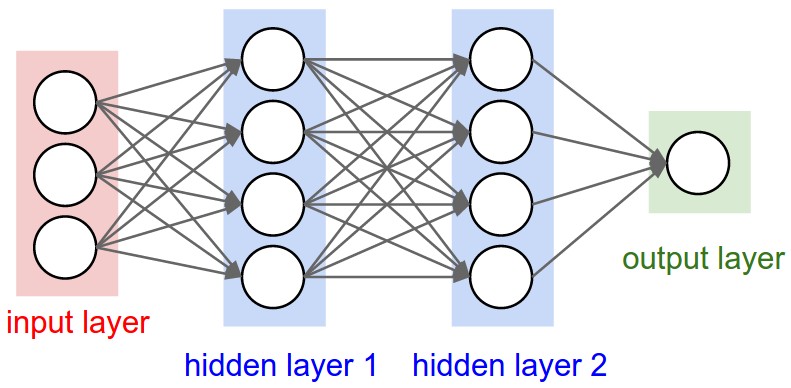
Figure 1. Illustration of an ANN.
In this example, the layers of the model are connected by mathematical functions akin to neurons (represented by circles). The neurons are developed during training and later ‘fired’ according to the data fed to them during testing. [1]
The research also involved the use of a completely different algorithm. Consider the 2-D space in Figure 2. The space already contained the solid red and blue points: the points on which the algorithm trains. The algorithm began by drawing random lines in the space between the red and blue points. These lines bisected the red and blue clusters such that all red points lay on one side of the line and all blue points on the other. For every such line drawn, the computer calculated the sum of the distance between the line and the closest red dot and the line and the closest blue dot. This distance was then maximized using calculus to give the maximum margin. And so a unique line was obtained with the maximum margin: the optimal hyperplane. Finally, when a prediction was to be made, the unknown point was plotted and depending on which side of the line (i.e. the optimal hyperplane) it falls, it was assigned a label- blue or red. A similar concept extends to higher dimensions in an algorithm called a support vector machine (SVM). This algorithm was used in my research.
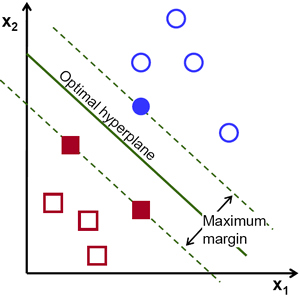
Figure 2. Illustration of a SVM.
Here, the optimal hyper-plane was identified by finding the line that had the largest distance from the 2 points closest to it, 1 in the red and blue class each. The hollow data points were then plotted and depending on which half-plane the points were in, they were classified as red or blue. The number of points required to determine the margin varies with dimensions. Here, 2 points were enough to determine a unique line. In 3D, 3 points determine a unique plane and so on. [2]
I also intend to employ these and several such algorithms to the project at hand at the Brindley lab. The algorithms described were devised in the 1950s; however, it is only in the last 15 to 20 years that the hardware has caught up, allowing researchers like me to put them in use. However, simpler and computationally less demanding algorithms do exists, such as the k-Nearest Neighbours algorithm and the logistic regression. I intend to apply the full spectrum of classification algorithms, from the simplest and intuitive to the most computationally demanding and mathematically rigorous, to the data in an effort to find the best performing one.
Predicting the Probability of Drug Launch: A Machine Learning-Based Approach
Historically, the pharmaceutical industry’s mode of operation is to rely on blockbuster drugs by conducting expensive clinical trials followed by marketing in developed economies if the trials are successful. Unfortunately, this tactic isn’t working very well. According to a recent report by the Boston Consulting Group, the cost of development of a single drug is often in the neighbourhood of 800 million USD [3].The drug attrition rate is 10-15% from the time a drug is first conceived as a potential medicine to its production for market use. That means that only 15 or so new molecular entities (NMEs) out of every 100 clear clinical trials [4].
My research project in the Brindley Lab will attempt to use machine learning methods to improve this dismal statistic. How? Machine learning is a tool that helps us understand how several quantities are related to one another purely on the basis of the data provided. It challenges preconceived notions about factors that influence an output. And, in the field of drug development, this is what is needed given the abysmal attrition rate and burgeoning costs. The algorithms that I will employ will highlight the factors that influence the probability that a drug will clear clinical trials. I plan on predicting the launch or failure of new molecular entities using economic, chemical and physiochemical variables to create a model that takes a holistic approach with regards to which variables affect drug success.
A pilot study with a small number of drugs and indications has already established some statistical relevance of these parameters. The parameters studied did demonstrate predictive potential. The next step in the project that I will be performing is to investigate further those variables that are useful in prediction, discard ones that show lower predictive power and find ways to efficiently group them in order to produce the best performing algorithm without overfitting to the data (see figure 3). This is also called feature selection. The problem to be solved is one of binary classification: will a drug candidate be launched or will it fail? Thus, I hope to use machine learning to identify the drugs that show a high probability of clearing these trials, cutting costs to pharmaceutical companies, reducing government spending on healthcare, and saving billions of dollars of the patients’ money.
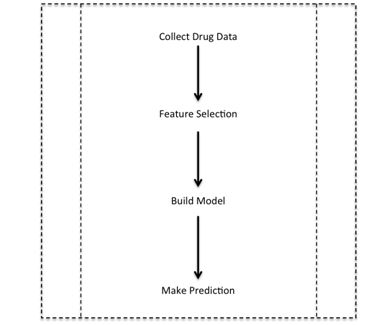
Figure 3. The steps in the project.
Step 1 involves the collection of a large amount of unbiased data. Step 2 consists of identifying the features that are relevant to prediction. Step 3 consists of actually building the model using the algorithms described, and Step 4 is the utilization of the model for future predictions.
Future Plans:
References:
[1] Stanford University Computer Science. CS231n, Convolutional Neural Networks for Visual Recognition. http://cs231n.github.io/
[2] OpenCV. Introduction to Support Vector Machines http://docs.opencv.org/2.4/doc/tutorials/ml/introduction_to_svm/introduction_to_svm.html
[3] Boston Consulting Group. A Revolution in R&D: How Genomics and Genetics are transforming the Biopharmaceutical Industry. Boston Consulting Group, Boston, Massachusetts, 2001. Page 6.
[4] Michael D Rawlins. Cutting the cost of drug development? Nature, April 2004, Volume 3, Pages 360-364.


