My name is Michaela Copp, and I am a rising senior studying Chemical Engineering at Vanderbilt University. While at Vanderbilt, I have been a member of the Neurovascular Engineering and Therapeutic Design lab under the direction of Dr. Ethan Lippmann. For the past two years, the Lippmann Lab has worked to develop the genetic engineering tools necessary to better model neurodegenerative disease pathogenesis and determine the individual genes responsible for the highly-specialized barrier properties of the blood-brain barrier (BBB).
The blood-brain barrier is responsible for regulating the transport of molecules between the bloodstream and brain to maintain proper neurological function. Although significant research effort has been placed on understanding the formation and regulation of the BBB, the distinct genes responsible for the resistance phenotype of the BBB remain relatively unknown. Fortunately, the discovery of the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9 system in 2012 has revolutionized researchers’ ability to interrogate gene function. The system consists of two key components: a Cas9 endonuclease responsible for cleaving DNA and a single guide RNA (sgRNA) that directs Cas9 to the gene of interest. The system can be engineered to target specific sequences in the human genome for editing by simply altering the nucleic acid sequence of the sgRNA [1]. To generate a gene knockout using the CRISPR system, the Cas9 protein and sgRNA must be delivered into the cell line. Although various methods of delivery have been used successfully in the past to achieve functional gene knockouts, the techniques generally have a low cutting efficiency in stem cells—between 5-10% knockout—or result in significant cell death [2]. The goal of my research project was to optimize a method of delivery of guide RNA into an induced pluripotent stem cell (iPSC) line to achieve an efficient gene removal, which can ultimately facilitate studies of BBB function in vitro.
The limitations of prior transfection methods led me to pursue an alternative method of delivery, Induced Transduction by Osmocytosis and Propanebetaine (iTOP) [3]. This transduction mechanism uses a hypertonic solution to trigger the nonselective uptake of extracellular molecules into cells. The concentration difference between the surrounding hypertonic solution and the interior of the cells drives the transport of solute molecules through the cell membrane. Analysis of immunostained images from experiments using the iTOP transduction method revealed gene knockouts greater than 60%, a vast improvement from traditional methods. Establishing an effective technique to edit a cell’s genome is a vital step in interrogating the function of genes. By removing particular genes within cultured cells that constitute a BBB model, our research lab can now assess their contribution to BBB function.
Use of the CRISPR System to Generate a Safe Harbor Landing Site for the Allotopic Expression of Mitochondrial Genes
This summer, I will be working with the SRF Mitochondrial Team under the guidance of Dr. O’Connor and Dr. Boominathan. The objective of my research project is to establish a safe harbor landing site – a position in the genome capable of integrating new genetic material without posing a risk to the host cell– in the nucleus for the expression of engineered mitochondrial genes. Mitochondria generate the cellular energy consumed by mammalian cells through the process of oxidative phosphorylation. Like the nucleus, mitochondria possess their own DNA, termed mtDNA, which encode for 13 proteins critical to cellular respiration. Unfortunately, mitochondria do not have an efficient system for repairing damaged DNA, leading to mutation rates 10 times greater than that detected in nuclear DNA [4]. Scientists believe evolutionary forces have driven mitochondrial genes from the mitochondria into the nucleus, where they are protected from the highly-reactive oxygen molecules produced by oxidative phosphorylation. The SRF Mitochondrial team hopes to mimic this evolutionary process by providing cells with a modified “backup” copy of the remaining mitochondrial genes at a safe harbor within the nucleus. The procedure of expressing genes in the nucleus originating from the mitochondria is called allotopic expression.
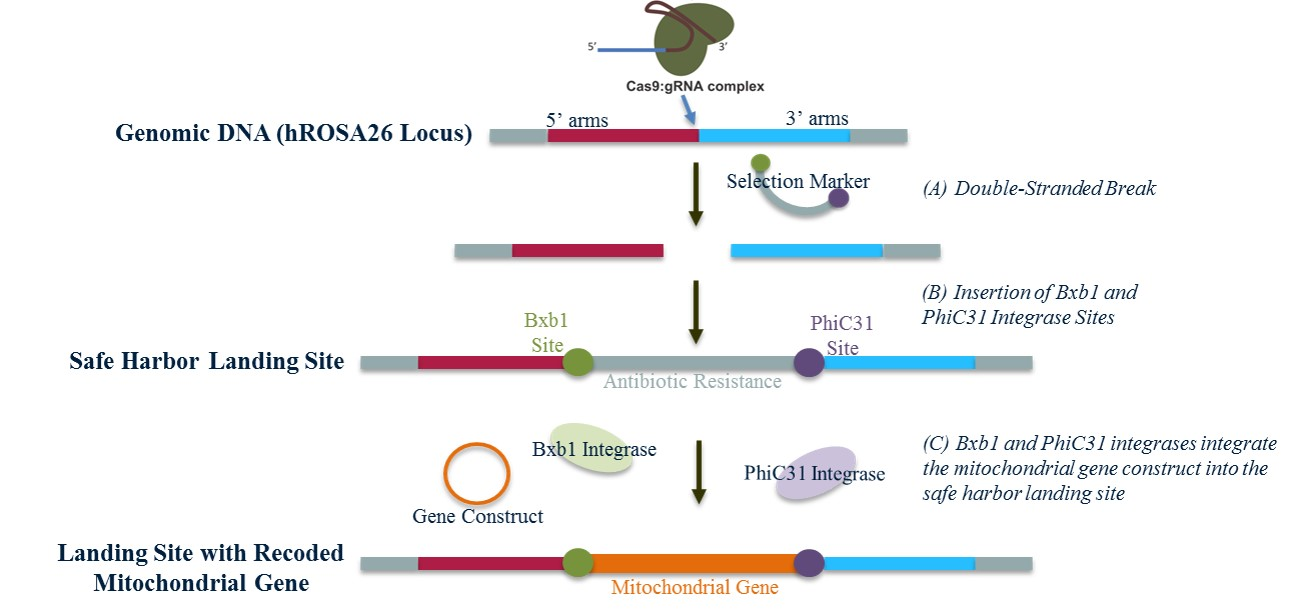
Figure 1. Generation of a Safe Harbor Landing Site and Expression of Recoded Mitochondrial Genes.
(A) Guide RNA’s targeting the hROSA26 locus form a complex with Cas9 to create a double-stranded break in the genomic DNA at the hROSA26 locus. (B) Bxb1 and PhiC31 integrase sites, flanked by hROSA26 homologous sequences, are inserted into the hROSA26 locus through homologous recombination. (C) From the landing site, recoded mitochondrial gene constructs may be integrated into the hROSA26 locus with the use of the Bxb1 and PhiC31 integrases.
Prior to this project, allotopic expression studies on mitochondrial genes had been performed via traditional transfection / virus induction procedures which integrate the new DNA randomly into the host genome. The goal of this study is to express the mitochondrial genes from an identified safe-harbor site in the nucleus, the hROSA26 locus, in order to minimally disrupt the host genome and ensure the gene functions predictably [5]. This will be accomplished by targeting the hROSA26 locus using the CRISPR system, creating a double-stranded break, and introducing a new strand of DNA containing a selection marker flanked by the attp sites of two different integrases (in this case Bxb1 and PhiC31) on either ends. These attp sites will function as “landing sites” to introduce different allotopic constructs. Integrases catalyze the site-specific integration of foreign DNA into a chromosome, and the dual integrase system will allow us to insert our mitochondrial genes with a directionality and specificity of 100% [6]. A diagram of this procedure is outlined in Figure 1. The benefit of using this allotopic expression approach is that the integrase system has no size limitations and provides for the integration of the mitochondrial genes at a precise location in the genome. Ultimately, this project will allow the SRF Mitochondrial team to establish a safe harbor landing site with the capability of integrating the entire mitochondrial genome into the nucleus.
Future Plans:
Moving forward, I hope to continue the biotechnological research I have begun in my undergraduate years and pursue a PhD in Bioengineering to develop a better understanding of the pathogenesis of neurodegenerative diseases, such as Alzheimer’s, Parkinson’s and Huntington’s. As the population gets older, age-related neurodegenerative diseases will become more prevalent. Presently, most of these diseases remain incurable and most available therapies are ineffective and have significant side effects. My goal upon obtaining a doctorate in Bioengineering is to direct research and development for a biotechnology company and develop targeted therapies to better combat the onset of these neurodegenerative diseases for a rapidly aging population.
References:
[1] X. Liang, P. Jason, K. Shantanu, Y. Zou, R. Quintanilla, M. Sridharan, et al. “Rapid and Highly Efficient Mammalian Cell Engineering via Cas9 Protein Transfection.” Journal of Biotechnology 208 (2015): 44-53. Web.
[2] J. Doench, N. Fusi, M. Sullender, M. Hegde, E. W. Vaimberg, K. F. Donovan, et al. “Optimized SgRNA Design to Maximize Activity and Minimize Off-target Effects of CRISPR-Cas9.” Nature Biotechnology 34.2 (2016): 184-91. Web.
[3] D. D’Astolfo, R. J. Pagliero, A. Pras, W. R. Karthaus, H. Clevers, V. Prasad, et al. “Efficient Intracellular Delivery of Native Proteins.” Cell 161.3 (2015): 674-90. Web.
[4] C. Haag-Liautard, N. Coffey, D. Houle, M. Lynch, B. Charlesworth, and P. D. Keightley. “Direct Estimation of the Mitochondrial DNA Mutation Rate in Drosophila Melanogaster.” PLoS Biology 6.8 (2008): n. pag. Web
[5] E. P. Papapetrou, and A. Schambach. “Gene Insertion into Genomic Safe Harbors for Human Gene Therapy.” Molecular Therapy 24.4 (2016): 678-84. Web.
[6] F. Zhu, M. Gamboa, A. P. Farruggio, S. Hippenmeyer, B. Tasic, B. Schule, et al. “DICE, an Efficient System for Iterative Genomic Editing in Human Pluripotent Stem Cells.” Nucleic Acids Research 42.5 (2013): n. pag. Web.


