My name is Julia McCreary, and I am a rising senior at University of California-Berkeley. I am a Molecular and Cell Biology major with an emphasis in biochemistry. From an early age, I’ve had a passion for research and have always enjoyed my time in the laboratory. I am still trying to find the “right” field of research but have narrowed my interests to the medical field and drug development. I was extremely interested in the research opportunities through the SRF Summer Scholars program because of their emphasis on disease and aging-related problems.
Most recently, I participated in research through UC Berkeley’s Museum of Vertebrate Zoology. Working in Rauri Bowie’s laboratory, I examined population genetics of avian species in Africa to understand evolutionary patterns between populations of birds. Comparing sequences, especially on highly conserved genes, is an important method to analyze relatedness between two individuals/populations. Sequences that are more similar imply a closer level of relatedness, while differences may be indicative of an adaptation that occurred between the two individuals. Our lab specifically looks for single nucleotide polymorphisms, or SNPs, which are a form of genetic variation in which individuals vary by a nucleotide at a specific base pair location. SNPs are important because they are the primary form of genetic variation among a species and can be used to study gene flow, or the movement of genetic material from one population to another. Gene flow is crucial to understanding evolutionary relationships and studying it can reveal patterns of immigration/emigration and the introduction of new alleles into a gene pool. My project is utilizing DNA extraction and sequencing to understand the evolutionary link between the Shelley’s greenbul and the stripe-faced greenbul.
Regulation of the proteostasis checkpoint in Drosophila intestinal stem cells
This summer, I am working in Heinrich Jasper’s lab at the Buck Institute for Research on Aging under the mentorship of postdoctoral fellow Imilce Rodriguez-Fernandez. The goal of my project is (1) to screen for additional candidate genes involved in regulating a novel mechanism used by adult somatic stem cells to deal with protein aggregates termed as the ‘proteostasis checkpoint’ and (2) to generate genetic tools to further understand protein homeostasis in these stem cells.
My project will help examine one of the hallmarks of aging, which is the loss of protein homeostasis (also known as proteostasis). Proteostasis refers to all the biological pathways involved in the synthesis, folding and degradation of proteins within a cell. As cells age, the mechanisms and cellular pathways by which they regulate the removal of misfolded/aggregated proteins deteriorates and non-functional proteins accumulate in the cells. Loss of proteostasis results in impaired cell function and contributes to a range of diseases such as Huntington’s, Alzheimer’s and Parkinson’s disease.[1]
Stem cells found in regenerative tissues, such as the intestine, are particularly sensitive to the loss of proteostasis. Intestinal stem cells are needed to maintain healthy tissues throughout the organism’s life. Using the Drosophila intestinal stem cells (ISCs) as a model system, the Jasper lab recently identified the ‘proteostasis checkpoint’ as a novel molecular mechanism used by these types of stem cells to deal with protein aggregates. Young ISCs are able to eliminate protein aggregates by undergoing a halt in the cell cycle until aggregates are removed. Older ISCs have more trouble disposing of protein aggregates than younger cells. Furthermore, because older ISCs cannot activate the ‘proteostasis checkpoint’ they continue to divide in the presence of aggregates.[2]
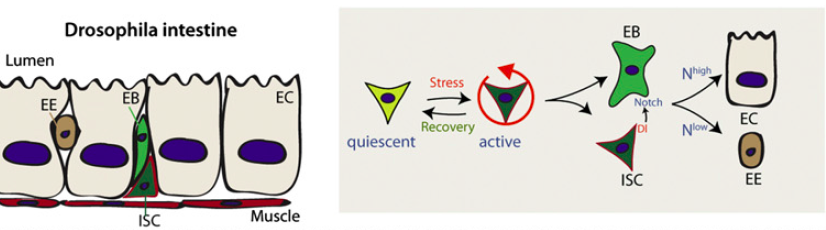
Figure 1. A schematic of the Drosophila intestine and the different cell types.
Intestinal stem cells (ISC) are the only mitotic cells in the intestine and give rise to EnteroBlasts (EBs), which then differentiate into EnteroEndocrine cells (EEs), the secretory cells of the Drosophila intestine, and EnteroCytes (ECs), the absorptive cells.[2]
The goal of my first project is to determine which genes regulate this checkpoint in young ISCs. To screen for new candidates we are using the GAL4/UAS system in flies, which allow us to express fluorescently-tagged aggregates in a temporally-controlled manner. GAL4 is specifically driven in ISCs and daughter cells known as EnteroBlasts (EB) using the escargot (esg) promoter. Transgenes under the UAS control will be expressed only in the cells expressing GAL4 (i.e. ISCs and EBs). Moreover the expression of GAL4 will be controlled by a temperature-sensitive version of its inhibitor known as GAL80ts. When flies are placed at a higher temperature, GAL80ts is nonfunctional and GAL4 can activate expression of the transgenes.
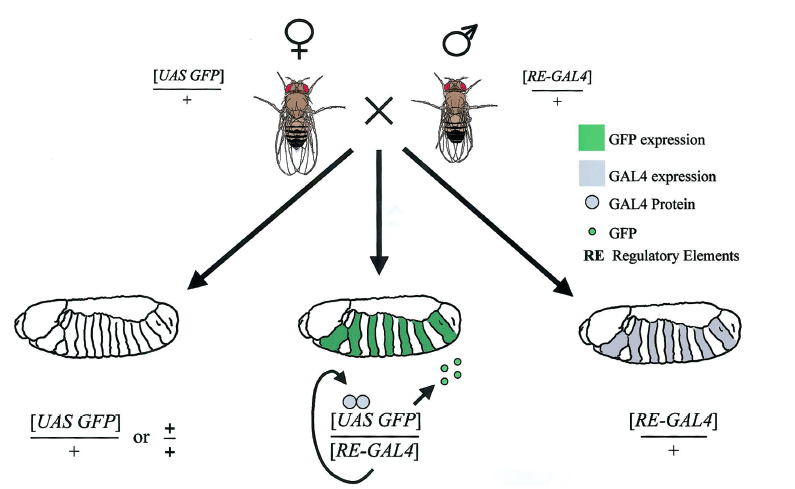
Figure 2. The UAS/GAL4 system in flies.
A male fly with the GAL4 gene is mated with a female fly with a UAS GFP gene. When offspring inherit both desired alleles, GAL4 is expressed and activates the transcription of GFP, green fluorescent protein, which allows us to track expression and determine which offspring have the desired genotype. A similar system is used in our project.[3]
To generate aggregates in ISCs and EBs, flies will carry a transgene encoding a fluorescently-labeled pathogenic version of human Huntingtin (Htt) with 138 polyglutamine repeats (mRFP-HttQ138) (generated by Weiss et al. 2001)[4]. The expression of mRFP-HttQ138 leads to the formation of aggregates that can be followed in the cells by fluorescence, which makes this transgene a great tool to study the elimination of aggregates.
The Jasper lab found that flies carrying aggregates in their ISCs have a higher mortality when treated with the oxidative agent Paraquat, a chemical that damages the intestine and activates regeneration of the gut. Because aggregates activate the ‘proteostasis checkpoint’ in ISCs, cells with aggregates failed to divide even when the gut was damaged and needed to be regenerated, leading to death of the animal.[2] This experimental paradigm will be the basis of my screening. RNA interference (RNAi) is a process in which small RNA molecules silence genes usually by destroying mRNA molecules so proteins are not formed. Using Gal4-dependent RNAi lines I will screen for those genes that ‘rescue’ the lethality observed when ISCs express aggregates.
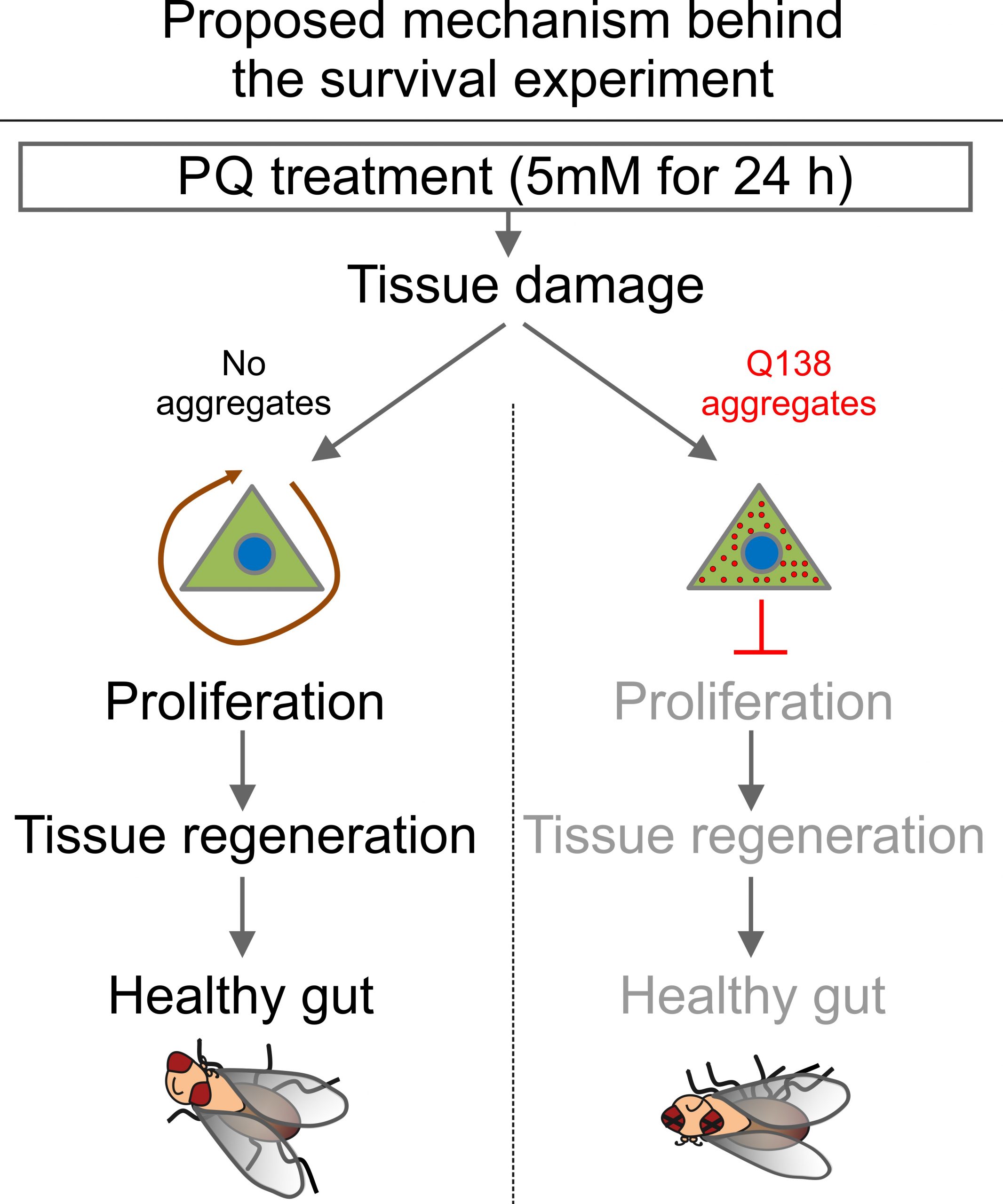
Figure 3. Proposed mechanism behind the survival experiment.
Under stressful conditions (treatment with Paraquat) ISCs with aggregates experience a halt in the cell cycle and are unable to rejuvenate damaged intestinal cells, leading to a higher mortality rate. We will screen for genes that are involved in the proteostasis checkpoint by identifying ones that yield a lower mortality rate when knocked down.
The goal of my second project is to generate two transgenes that will be used as tools to understand better how ISCs maintain their proteostasis. Using molecular biology techniques, I will create these transgenes to then generate transgenic flies.
Future Plans:
I plan on remaining in the Bowie lab to complete my senior thesis. After graduating from UC Berkeley, I hope to continue my research career outside of academia and work for a company for a year or two. I then intend to apply for a PhD/PharmD program to pursue my passion for pharmaceutical research.
References:
[1] Serrano, M.,Lopez-Otin, C., Blasco, M., Partridge, L., and Kroemer, G. (2013). The hallmarks of aging. Cell 153, 1194-1217.
[2] Jasper, H., Biteau, B., and Hochmuth, C. E. (2011). Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 9, 402-411.
[3] Duffy, J. B. (2002). GAL4 System in Drosophila: A Fly Geneticist’s Swiss Army Knife. Genesis 34, 1-15.
[4] Weiss, K., Kimura, Y., Lee W.C., and Littleton, J. (2012). Huntingtin Aggregation Kinetics and Their Pathological Role in a Drosophila Huntington’s Disease Model. Genetics 190, 581–600.


