I have just completed my third year of a Natural Sciences degree at Cambridge University in the United Kingdom. This year, I specialised in biochemistry, but the beauty of this major is that it allows me to study a wide range of subjects. In my first year, I studied math, chemistry and materials science, while I studied plant science and developmental biology in my second year. I have decided to focus on biochemistry for my fourth and final year, as I think the current advances in physical and biological sciences together will stimulate the next breakthroughs in modern healthcare via biomedical innovation.
Last summer, I undertook a computational data analysis research project at the Institute of Cancer Research in London. Understanding how cancer cells respond to different drugs is crucial to developing effective chemotherapy. However, most current drug screens primarily usescreen for very simple readouts, such as cell death or a change in concentration of a target molecule (Hoelder et al., 2012). Dr. Chris Bakal’s lab is developing new ways of investigating the effects of cancer drugs, based on high-throughput measurement of hundreds of cell morphology parameters. Screening cell morphology changes should is thought to give give a broader indication of the multifaceted effects of many candidate cancer drugs, but extensive data analysis is required to interpret the large amounts of morphology data.
To help address this problem, I implemented various algorithms to cluster cancer cell morphology data. These algorithms basically tell the computer to sort all available data into distinct groups, or clusters, based on similarity. Similar clustering algorithms are used every day to recognise communities in online social networks. In Figure 1, you can see that data clustering allowed me to identify 5 distinct morphological “outcomes” that are caused by different anti-cancer drugs. In this study, I was investigating the effect of drugs that are thought to antagonise key proteins involved in the regulation of cell shape, such as those that regulate the cell “skeleton,” which is composed of polymers of millions of individual actin subunits. The three most extreme departures from the “normal” HeLa cell shape are labelled with representative images. Although only a few morphological differences between can be discerned by the naked eye, a computer can detect many subtler differences between the images, some of which may be biologically relevant.
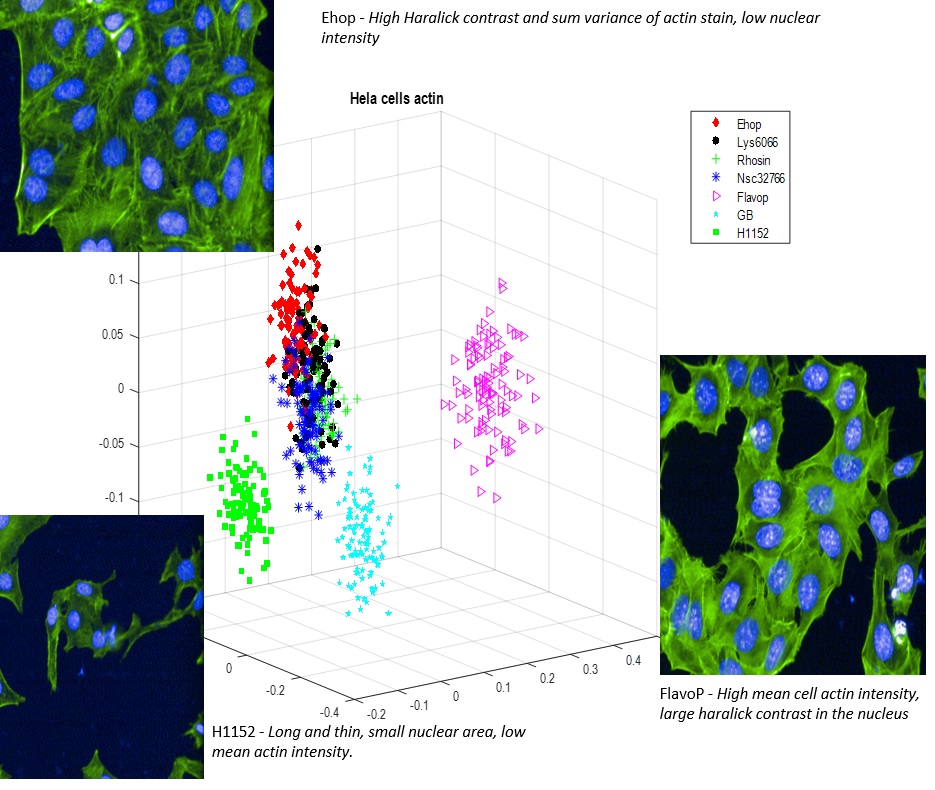
Figure 1. Cancer cell morphology data clustered into distinct groups based on similarity.
This graph depicts a small subset of cells analysed, whose 63 morphology parameters have been transformed into just 3 dimensions (for ease of interpretation). Data clustering shows that drug-specific effects on these cancer cells can be clearly detected. The different shapes and colours correspond to different drug treatments, as shown by the legend. Flavopiridol, H1152 and Ehop drug treatments produced the most dramatic effects on cell morphology, and the differences between these 3 effects are illustrated with a representative image of each treatment. Cellular actin is imaged by fluorescently labelling phalloidin (in green), and the cell nuclei are labelled with DAPI (in blue).
Early prediction of small molecule market authorization: a machine learning approach
Age-related diseases affect most people on Earth, and their complex, multifaceted nature makes repairing the damage caused by these diseases a particular challenge. A significant proportion of global healthcare budgets are spent on alleviating the symptoms of age-related disease, and the rapidly expanding and ageing world population will only amplify these issues.
There is, therefore, a pressing demand to develop novel chemotherapeutics that can meet this challenge, however advancements in drug discovery are currently limited by the development process. Drugs that show promise in an in vitro model system must first demonstrate a beneficial effect in animal models, then pass through three phases of human clinical trials to be deemed successful (Figure 2). This process takes on average ten years and can be prohibitively expensive. Furthermore, 85% of pre-clinically approved drugs fail in clinical trials (Mak et al., 2014), and, of those that pass all 3 phases and have their New Drug Application approved (Figure 2), various compounding factors often further increase the time taken to reach the market. Time and money are finite resources, and, considering the high failure rate of the current drug discovery process, it is highly desirable to economise these resources and ensure they are spent effectively.
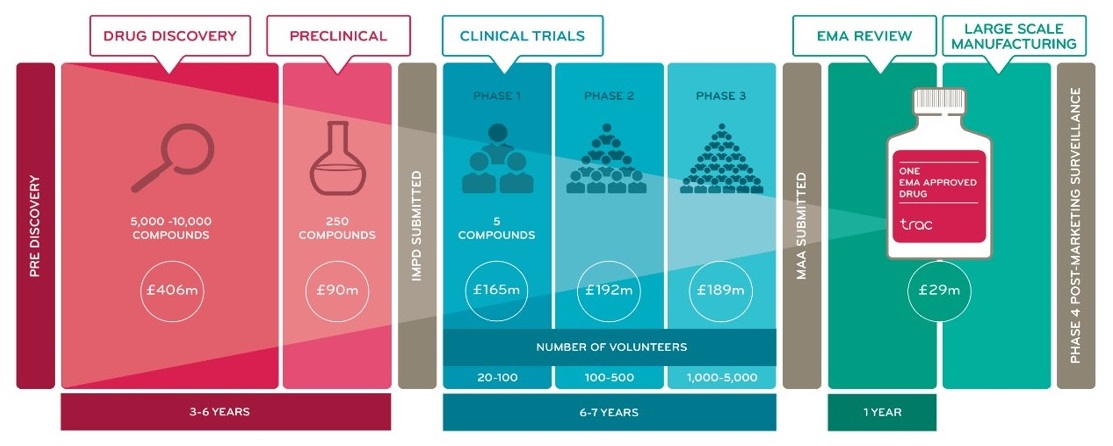
Figure 2. Timeline of a typical drug development process, with representative figures.
This figure refers to the drug development process in the United Kingdom, but the timeline and regulatory steps are similar worldwide. IMPD stands for an Investigational Medicinal Product Dossier, which certifies approval for clinical trials to take place; the US equivalent is the INDA (Investigational New Drug Application). A Marketing Authorisation Application (MAA) must then be approved at the end of clinical trials, before the drug can be commercially distributed; this is equivalent to the NDA (New Drug Application) in the United States. In Europe, the European Medicines Agency (EMA) then further reviews the commercialised product, as does the FDA in the US.
Image taken from Trac Services: https://www.tracservices.co.uk/blog/trac-services-drug-development-process/
As a SRF Summer Scholar, I will be investigating a diverse array of data pertaining to previously conducted clinical trials in search of variables that frequently correlate with success. Identifying these so called ‘predictor variables’ may give us a better indication of which future trials are likely to be successful and, in the long run, may minimize time and money waste by reducing the high failure rate of therapeutic development. I will collect a broad range of input data for this task, from drug activity indications to company attributes, patents, and even macroeconomic trends during the drug discovery process, in an attempt to identify which of these input variables are frequently linked to “success.”
However, the ultimate “success” of a drug is defined by a variety of outcomes, all of which should be considered equally at the start of this discovery process. Moreover, the eventual identification of predictors will depend on the specific outcome variable chosen to be indicative of success. For this project, I will define the outcome variable as approval of the Mmarket Aauthorisation Aapplication (MAA, see Figure 2). However, there are numerous other factors that subsequently influence the time taken to commercially distribute the approved drug and the eventual success of the drug as a commercial product. Therefore, I will also investigate what factors influence these subsequent steps and whether predictor variables can be reliably established at these steps.
Identifying reliable predictor variables requires computational power, which vastly increases the amount of data that can be analysed in a given time period. Particularly applicable to this task are some high-throughput computational data analysis algorithms that all come under the umbrella term of ‘machine learning.’. Simply put, machine learning algorithms allow a computer to “learn” and subsequently predict outcome variables of interest, without being explicitly programmed in advance.
For example, say I want to predict the cost of my house in 10 years. One way I could do this is to collect data detailing the changes in house prices near my address for the last 50 or 100 years. Having done this, I could plot the input variable (year) against the outcome variable (house price), as below (Figure 3).
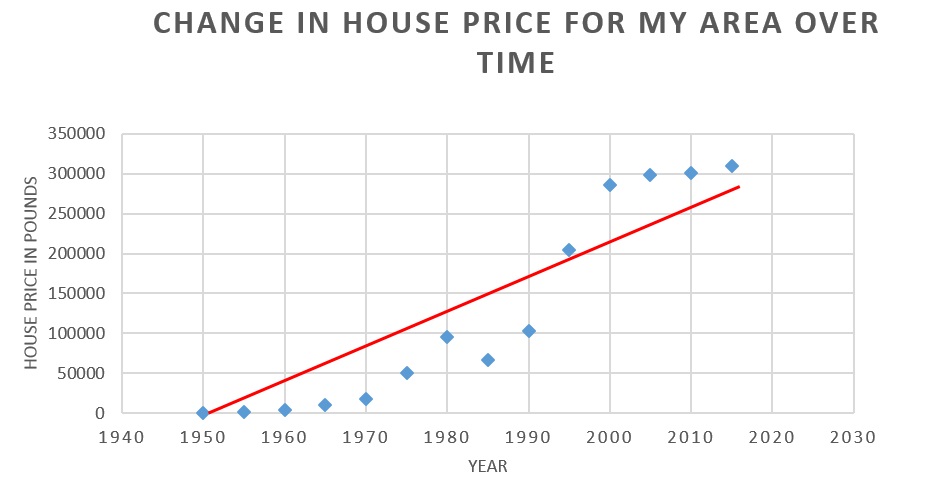
Figure 3. A real-world example of linear regression.
This example graphs the change in house price over time. The red line represent the best fit function for the data.
Now, to predict my house price in 2026, I could try to fit a line to this data and extrapolate (e.g. red line above). Importantly, the predicted outcome variable will depend on the particular line fitted. This line can be described by a function h(x), where h is the set of operations that are performed on the input variable(s) X to get the outcome variable.
To improve the accuracy of prediction of the fitted line, we want its value at each point to be as close as possible to the data provided. This can be achieved by measuring how close the match is at each point (using a “cost function”), then drawing a new line that is a closer match. Supervised machine learning algorithms simply iterate this process, so that eventually a function h(x) is found which best describes the data, and can, therefore, most accurately predict my house price in ten years’ time.
In this example, all data regarding local house prices in previous years is referred to as the “training set”, as it is used to improve or “train” h(x) to eventually best predict future house prices. Figure 4 illustrates the basic process illustrated here, which is used in all supervised machine learning:
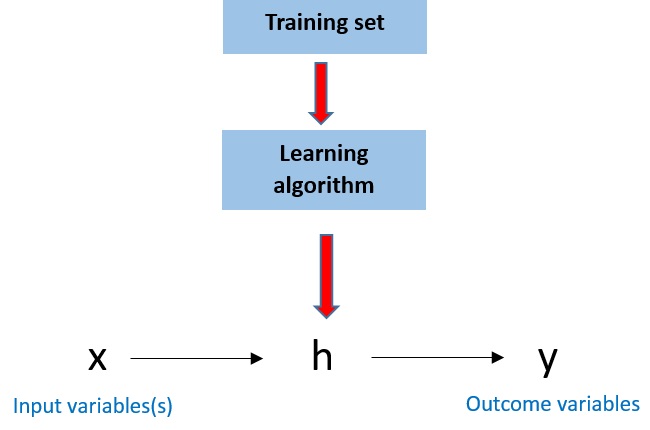
Figure 4. A simple flowchart illustrating the process used in supervised machine learning.
By first “training” your algorithm on data for which you know the outcome of interest, you can then use it to predict the outcome of interest in new data.
The supervised machine learning described above for house prices is in many ways analogous to the way it will be employed to investigate predictor variables of success in drug development. In my project, X will represent a variety of input variables, whilst Y represents market approval of a drug. In order to obtain the most accurate prediction of future house price, it would be useful to collect more data on a wider range of variables: it seems plausible that many other compounding factors would also influence local house price, such as economic or political changes or the companies involved in selling the houses. The same is true for the success of a new drug: a wide range of variables is likely play a role in ultimate success and incorporating as many of these input variables as possible into the training set maximises chances of being able to make accurate predictions.
Future Plans:
I will be staying in Cambridge to complete a masters in biochemistry next year, before pursuing a PhD. I am particularly interested in computational biology and its many applications in healthcare research. I hope to find myself working on similar projects to this one as a post- graduate researcher.
References:
1. Hoelder S, Clarke P & Workman P (2012) Discovery of small molecule cancer drugs: Successes, challenges and opportunities. Molecular Oncology 6: 155-176.
2. Mak I W, Evaniew N, & Ghert M (2014) Lost in translation: animal models and clinical trials in cancer treatment. Am. J. Transl. Res. 6: 114–118


