My name is Elizabeth Batiuk, and I am a rising senior studying bioengineering at Santa Clara University. During my time at Santa Clara University, I have been fortunate to work in Dr. Zhiwen (Jonathan) Zhang’s bioengineering laboratory. His laboratory focuses on “bioengineering towards medicine” and uses protein engineering to study mechanisms that lead to drug discovery.
Dr. Zhang’s laboratory had previously designed a tetracycline repressor-based mammalian two-hybrid (trM2H) system to monitor protein-protein interactions. Although there are a handful of methods to study protein-protein interactions that currently exist, such as the enzyme-linked immunosorbent assay (ELISA) or the yeast two-hybrid system, they are not as accurate as the trM2H system. ELISA quantifies and detects proteins, antibodies and peptides. However, since it is a plate-based in vitro assay, it cannot provide information about the interactions of these proteins in their natural environments. Another popular model for studying protein interactions is the yeast two-hybrid system, which can detect the physical binding between proteins. However, researchers are still unsure whether mammalian protein behavior in this system is representative of their natural environment. The Zhang lab’s novel trM2H system allows researchers to more comprehensively study protein-protein interactions.1
My project in the Zhang lab is focused on the interaction between Jab1, a signaling protein, and psoriasin, a growth factor receptor. Both of these proteins are involved in estrogen receptor alpha (ERα) breast cancer, but it is still unclear whether they interact with each other.2 Binding strength information would verify the interactions of these proteins and may suggest a more detailed causal mechanism. Prior in vivo studies in Zhang’s lab using the trM2H system have also suggested the ability of both of these proteins to form homodimers (compounds composed of two of the same protein) as well as heterodimers (compounds composed of one of each protein). Data correlating the in vivo studies with the actual binding affinity of Jab1 and psoriasin can further support the trM2H system’s data and help it more accurately capture the binding affinity of thousands of protein-protein interactions in the future. Thus far, my role in this project has involved cloning the genes and purifying the resulting proteins by affinity chromatography. The proteins’ binding strength to themselves and each other will be determined using Isothermal Titration Calorimetry (ITC) to record the exact energy change of the proteins.
Any information regarding the protein interaction of Jab1 and psoriasin would be useful to better understand their roles in ERα breast cancer and could potentially lead to the development of therapeutic drugs. If a correlation between binding strength of these proteins and the stage of ERα breast cancer can be determined, the interaction between these proteins can also be used as a diagnostic tool. Additionally, this research will be essential in confirming the Zhang lab’s work with the mammalian trM2H system.
I am excited to further develop my laboratory skills with the SENS Research Foundation Summer Scholars Program this summer. I am very interested in their approach of preventing the onset of age-related diseases rather than treating the side effects later. I look forward to learning new laboratory techniques and gaining insight on how to conduct scientific research from the many knowledgeable scientists at the SRF Research Center.
Allotopic expression of mitochondrial gene ATP8 to rescue function in mouse cells
This summer, I will be joining the Mitochondria Team, led by Dr. Matthew O’Connor at the SRF Research Center. Mitochondria produce the majority of chemical energy for mammalian cells through cellular respiration. Besides the nucleus, the mitochondrion is the only other organelle in mammalian cells that possess its own DNA. However, unlike the nucleus, mitochondria lack effective DNA repair mechanisms, which results in mutation rates that are 10 times higher than those observed in nuclear DNA.3,4 Over the course of human evolution, scientists believe the majority of mitochondrial genes have transferred to the nucleus. The proteins for which these genes code for are transcribed, translated and targeted to the mitochondria. Since the nucleus has better repair mechanisms, it is less likely that these transferred mitochondrial genes will mutate. Mitochondrial DNA now consists of only 13 proteins. Mutations in the remaining genes in the mitochondrial genome can lead to blindness, neurological, cardiac, respiratory and gastrointestinal diseases and have also been found in cancerous tissues.5 Approximately 9 in 100,000 people have mutations in their mitochondrial DNA.6 1 in 200 children inherit genetic mutations in mitochondrial genes each year and experience these symptoms as well.7
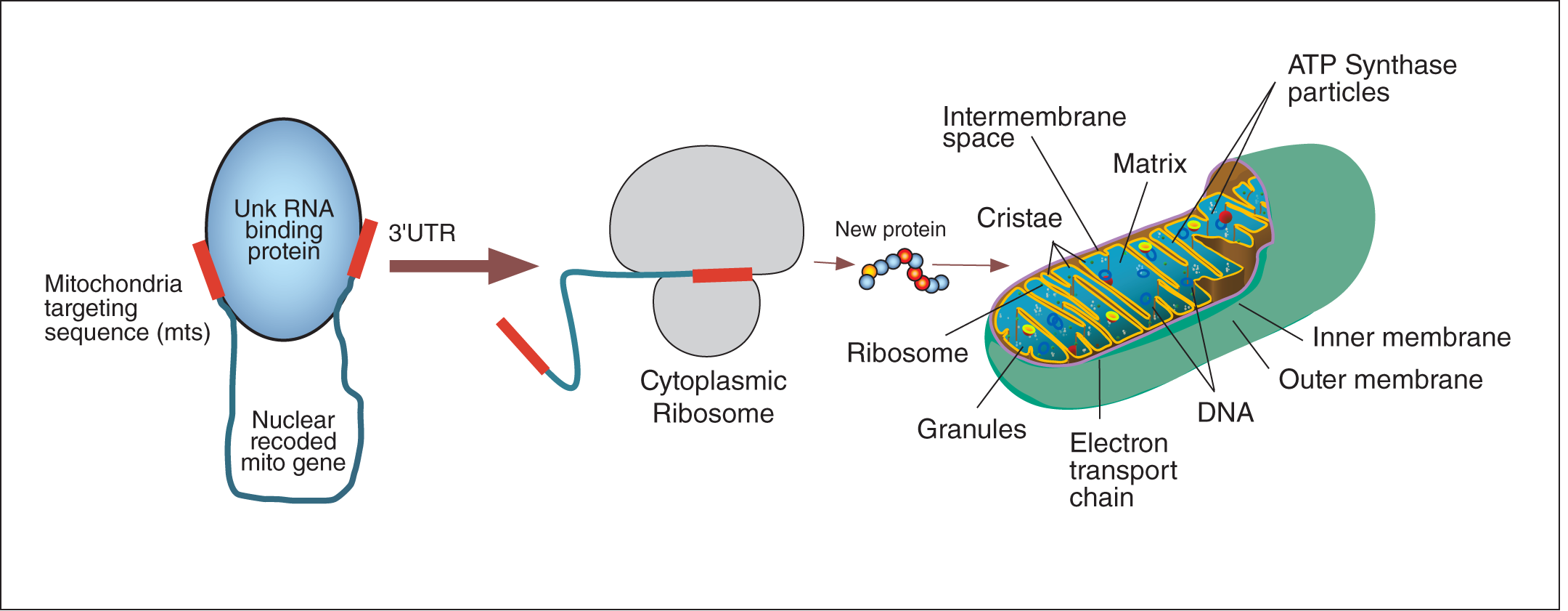
Figure 1. Allotopic Expression.
The nuclear encoded mitochondrial gene is transcribed in the nucleus, translated in the cytoplasm, and targeted and imported to the mitochondria with the help of the mitochondria targeting sequence to restore function.
An approach to solving this problem is to look at the pre-existing biological solution and allotopically express the non-mutated mitochondrial genes. I will obtain a cell line with a mutation in a certain mitochondrial gene, integrate a non-mutated version of the gene into the nuclear genome, and determine whether the function of that gene’s protein has been restored by testing for cell viability and energy production. In particular, my project will focus on rescuing the function of a protein in a mouse cell line with a mutation in the mitochondrial gene, ATP8, an important subunit of mitochondrial complex V, which is essential to producing the chemical energy for the body. I will start by determining the effect of the mutation on protein function. To rescue the aberrant protein function, I will integrate the non-mutated gene into the nuclear genome. In addition to verifying that the protein is correctly folded and directed to the mitochondria, I will also test viability, energy production capability, and measure the energy content to determine if protein function has been restored.
Future Plans:
In the fall, I will return to Santa Clara University where I will start my senior design project and complete the final year of my undergraduate degree. After graduation, I plan on taking a gap year while I apply to MD and MD/PhD programs. During this time, I hope to continue performing research in a laboratory as well as travel.
References:
1. Thibodeaux GN, Cowmeadow R, Umeda A, Zhang Z. A Tetracycline Repressor-based Mammalian Two-hybrid System to Detect Protein-protein Interactions in vivo. Analytical biochemistry. 2009; 386(1):129-131.
2. Emberley ED, Niu Y, Leygue E, Tomes L, Gietz RD, Murphy LC, et al. Psoriasin interacts with Jab1 and influences breast cancer progression. Cancer Res. 2003;63:1954–61.
3. Haag-Liautard C, Coffey N, Houle D, Lynch M, Charlesworth B, Keightley PD. Direct Estimation of the Mitochondrial DNA Mutation Rate in Drosophila melanogaster. Hurst LD, ed. PLoS Biology. 2008; 6(8)
4. Galtier N, Enard D, Radondy Y, Bazin E, Belkhir K. Mutation hot spots in mammalian mitochondrial DNA. Genome Res. 2006; 16:215-222.
5. Taylor RW, and DM Turnbull. “Mitochondrial DNA Mutations in Human Disease.” Nature Reviews. Genetics. U.S. National Library of Medicine, May 2005.
6. Schaefer AM, McFarland R, Blakely EL, He L., Whittaker RG, Taylor RW, Chinnery PF, Turnbull DM. Prevalence of mitochondrial DNA disease in adults. Ann Neurol. 2008; 63: 35- 39.
7. Elliott HR, Samuels DC, Eden JA, Relton CL, Chinnery PF. 2008; Pathogenic mitochondrial DNA mutations are common in the general population. Am J Hum Genet. 2008; 83: 254-260.


