My name is Le Zhang. I just completed my Bachelor of Science in biochemistry & biotechnology from Michigan State University. Before joining to the SRF Summer Scholars Program, I worked in Dr. Yonghui Zheng’s lab at MSU searching for novel proteins or molecules from our human body that can limit human immunodeficiency virus-1 (HIV-1) infection. These proteins are called restriction factors. One of the potential restriction factors we researched was the T-cell immunoglobulin and mucin domain (TIM) protein family. TIMs are T cell surface receptors which were reported in the past decade to promote envelope viruses (viruses surrounded by a lipid bilayer) binding and entry into the host cells like hepatitis A and Ebola1. However, one of the most recent studies has shown that TIM proteins 1, 3, and 4 strongly inhibit the release of HIV-1 2, but the study did not establish the mechanism by which viral proteins antagonize TIM proteins.
Under the supervision of Dr. Yonghui Zheng and Dr. Xianfeng Zhang, I focused on verifying if TIMs have the novel ability to inhibit HIV-1 and finding possible viral proteins that interact with TIMs. Our hypothesis was that HIV Negative Regulatory Factor (Nef), a viral protein which can down regulate host surface receptors like CD4, has the ability to also interact with TIMs. To test this hypothesis, I created an HIV-1 Nef knockout mutant in a retro-vector, which is a plasmid that encodes all HIV-1 proteins except for Nef and can assemble HIV particles. Three TIM plasmids were also prepared, which overexpressed TIM 1, 3 & 4 proteins, respectively. I co-transfected HIV-1 Nef knockout and TIMs plasmids in 293T cells, which are specially designed cells to overexpress plasmids. I determined the effect of overexpression of TIMs by measuring the amount of virus released by the Nef knockout strain compared to the non-mutant control. This research helped us better understand how the HIV infection mechanism works and how our immune system responds to the infection. It will serve as the foundation for future anti-viral treatment development in the Zheng lab.
I was attracted to stem cell research after learning about their power in tissue renewal, their utility in fighting aging, and other novel functions. I hope to learn more about stem cells during my internship with the SRF Summer Scholars Program.
Evaluating genomic stability of induced pluripotent stem cells for Parkinson’s disease cell therapy by SNP analysis
This summer, I will be conducting my research project in Dr. Jeanne Loring’s laboratory at the Center for Regenerative Medicine in the Scripps Research Institute. Guided by Dr. Michael Boland and Dr. Andres Bratt-Leal, my main goal for the summer is to analyze the genomic stability of induced pluripotent stem cells (iPSCs) and neuronal progenitor cells (NPCs), which are intended for cell replacement therapies for patients with Parkinson’s disease.
Human iPSCs are usually derived from somatic cells, like skin cells. By using biological or chemical methods, they are reprogrammed into stem cells. iPSCs are well known for their ability to self-renew and their power to differentiate into other cell types. These features make them a promising tool in cell transplantation therapy development. In many age-related diseases, irreversible cell death is often a major contributor. For instance, death of dopamine-generating cells, one type of neuron in the midbrain, leads to Parkinson’s disease. Previous research in rodent and primate models of Parkinson’s disease has shown that midbrain dopaminergic (DA) neurons differentiated from human embryotic stem (ES) cells improved forelimb use and movement control in Parkinsonian animals3. Also, ES cell-derived DA neurons showed long-term survival in animals3.
Use of human ES cells raises some morality concerns and can lead to resistance for use in adult treatments. However, use of iPSCs directly from patients addresses this concern and is more convenient and safer as well. Although iPSC research shows great promise, reprogramming has the potential to induce detrimental genomic changes. Expansion and differentiation of iPSCs over time could lead to genomic change and possibly tumorigenesis4. Past experiments from the Loring lab indicate that cell lines propagated over 100 continuous passages, regardless of passage methodology, experience genomic instability, such as genomic deletions and duplications5.
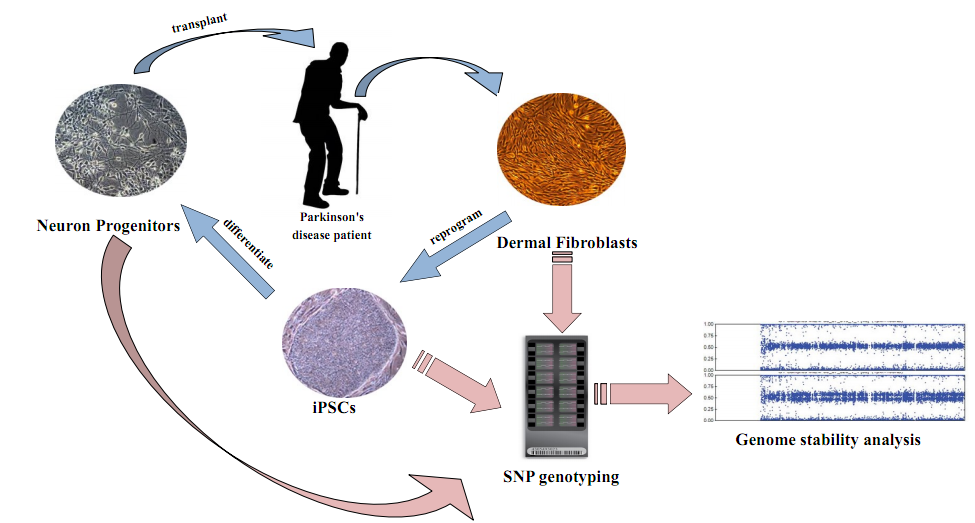
Figure 1. Parkinson's disease transplantation project overview.
The Loring lab has derived dermal fibroblasts from 10 patients with Parkinson’s disease. These fibroblasts have been reprogrammed to iPSCs, which have been differentiated into midbrain-specific NPCs. These cells will later develop into DA neurons after transplantation. The Loring lab is the first lab conducting iPSC transplantation on Parkinson’s disease patients, so it is essential to ensure genomic stability of the cells being transplanted. An important method to determine genomic integrity of patients’ iPSC lines is single nucleotide polymorphism (SNP) genotyping, which can be used to examine millions of single base pair differences at genomic sites specific to humans.
SNP analysis will enable me to determine if the cell populations are suitable for transplantation or whether they have too much genetic change and, hence, potential risk for tumorigenesis. My research this summer will generate and analyze genomic SNP profiles from patient-specific dermal fibroblasts, iPSCs, and neuronal progenitors. SNP patterns from the three cell types will be compared to determine whether genomic instability has occurred from fibroblasts to iPSCs then to neuronal progenitors. Hopefully, with efforts from other scientists and me, the Loring Lab will successfully identify some cell lines that are suitable for transplantation and pass the FDA approval.
Future Plans:
Stem cell research is new to me. Through my experiences in the Loring lab, I want to learn more and later contribute to this field. I plan to apply to graduate school after the SRF Summer Scholars Program has ended.
References:
1. Stephanie Jemielity, Jinyize J Wang, Ying Kai Chan, Asim A Ahmed, Wenhui Li, Sheena Monahan, Xia Bu, Michael Farzan, Gordon J Freeman, Dale T Umetsu, Rosemarie H Dekruyff, Hyeryun Choe. TIM-family Proteins Promote Infection of Multiple Enveloped Viruses through Virion-associated Phosphatidylserine. PLOS. Pathogens: 2013. e1003232.
2. Minghua Lia, Sherimay D. Ablanb, Chunhui Miaoa, Yi-Min Zhenga, Matthew S. Fullera, Paul D. Rennertc,Wendy Mauryd, Marc C. Johnsona, Eric O. Freedb,Shan-Lu Liua. TIM-family proteins inhibit HIV-1 release. PNAS: 2014, doi: 10.1073/pnas.1404851111
3. Sonja Kriks, Jae-Won Shim, Jinghua Piao, Yosif M. Ganat, Dustin R. Wakeman, Zhong Xie, Luis Carrillo-Reid, Gordon Auyeung, Chris Antonacci, Amanda Buch, Lichuan Yang, M. Flint Beal, D. James Surmeier, Jeffrey H. Kordower, Viviane Tabar & Lorenz Studer. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011. Vol 480, 547-551.
4. Fox JL FDA scrutinizes human stem cell therapies. Nat Biotechnol. 2008. Vol 26, 598-599.
5. Ibon Garitaonandia, Hadar Amir, Francesca Sesillo Boscolo, Geral K. Wambua, Heather L. Schultheisz, Karen Sabatini, Robert Morey, Shannon Waltz, Yu-Chieh Wang, Ha Tran, Trevor R. Leonardo, Kristopher Nazor ,Ileana Slavin , Candace Lynch , Yingchun Li , Ronald Coleman , Irene Gallego Romero ,Gulsah Altun , David Reynolds , Stephen Dalton , Mana Parast , Jeanne F. Loring & Louise C. Laurent. Increased Risk of Genetic and Epigenetic Instability in Human Embryonic Stem Cells Associated with Specific Culture Conditions. PLOS ONE. 2015. DOI:10.1371/journal.pone.0118307.
6. Louise C. Laurent, Igor Ulitsky, Ileana Slavin, Ha Tran, Andrew Schork, Robert Morey, Candace Lynch, Julie V. Harness, Sunray Lee, Maria J. Barrero, Sherman Ku, Marina Martynova, Ruslan Semechkin, Vasiliy Galat, Joel Gottesfeld, Juan Carlos Izpisua Belmonte, Chuck Murry, Hans S. Keirstead, Hyun-Sook Park, Uli Schmidt, Andrew L. Laslett, Franz-Josef Muller, Caroline M. Nievergelt, Ron Shamir, Jeanne F. Loring. Dynamic Changes in the Copy Number of Pluripotency and Cell Proliferation Genes in Human ES and iPS Cells during Reprogramming and Time in Culture. Cell Stem Cell. 2011. Vol 8. 106-118.


