My name is Karina Liker, and I am a rising senior at Stanford University. I am majoring in Human Biology with a concentration in Genetic Counseling and Diseases. I remember becoming interested in biology and regenerative medicine in high school when I learned about stem cell therapies. Since then, I have enjoyed learning about regenerative medicine and other clinical therapies in my biology coursework.
While attending Stanford, I have been fortunate to have been involved in a few projects in Professor Margaret Fuller’s lab in the Department of Developmental Biology. Last summer, I was involved in a project investigating how alternative polyadenylation and the length of 3’ untranslated regions (UTRs) affect mRNA expression in Drosophila (fruit fly) testis stem cells. A 3’ UTR is a sequence of DNA that is attached at one end of each strand of mRNA during transcription. At the end of the 3’ UTR is a poly(A) tail, which is a sequence of RNA adenine bases attached to the end of the mRNA before it is translated into a protein. Varying lengths of 3’ UTRs can affect the structure of the poly(A) tail since it is located on that same end of the mRNA; in other words, the mRNA can be alternatively polyadenylated. Moreover, different lengths of the 3’ UTR can have different implications for gene expression. For instance, an RNA-binding protein may be able to bind to one 3’ UTR sequence but not others and, in this way, block or increase mRNA expression.
To identify how alternative polyadenylation and 3’ UTR length affects development and gene expression in sperm stem cells, particular length 3’ UTRs of genes of interest will be expressed in different regions of the Drosophila testis. That will allow the effect of the different 3’ UTRs on sperm stem cell development to be observed through microscopy, immunofluorescence, and in situ hybridization. Last summer, I began the project by creating various recombinant plasmids each containing a particular 3’ UTR of a different gene of interest. The plasmids were then prepared for injection into Drosophila embryos. Although some of the embryos have been injected, the project is still underway, and I look forward to discovering the significance of polyadenylation in sperm stem cell fate.
Over the last few quarters, my research project in the Fuller Lab has been to identify new RNA-binding proteins that may affect sperm stem cell development by conducting an RNAi screen. Using specific lineages of genetically engineered flies, we can make certain regions of testis stem cells express short RNA sequences that bind specifically to mRNA segments. These particular strains encode sequences that bind to the mRNA of RNA-binding proteins and block expression of the mRNA. By restricting the knockdown of these genes to either the early-stage stem cells or later-stage stem cells, we can get an idea of whether the gene is necessary for sperm cell development throughout the entire process or only in a particular phase of spermatogenesis. So far, I’ve found several “hits,” or proteins that produced an unusual phenotype in the testis (usually loss of sperm) when mRNA expression was knocked down. Most of the hits I’ve observed are proteins involved in spliceosome function during transcription, and a few have human homologs.
I was initially interested in becoming a SRF Summer Scholar because I was curious about working in an environment focused on more clinical-type research studies and about working with mammalian research models. Under the mentorship of Dr. Ehud Goldin and the rest of the lysosomal research team, I will be exploring the biological basis of age-related macular degeneration (AMD) to eventually create a treatment for the disease.
Creating a Cell-Based Model of Age-Related Macular Degeneration
The macula is the central part of the retina located in the back of the eye. The photoreceptor cells it contains in one section, called the fovea, are responsible for most of our acute, clear vision. In the early stages of macular degeneration, lipids and other photoreceptor waste products accumulate beneath the photoreceptors in the underlying membranes, like the Bruch’s membrane and retinal pigment epithelium (RPE). The lipid deposits are called drusen, and large drusen seen through an opthalmoscope (an instrument used by ophthalmologists to view the inside, back surface of the eye) are used to diagnose early AMD. As the patient ages, early AMD can progress to geographic atrophy (“dry” AMD), in which photoreceptors die, or neovascular AMD (“wet” AMD), in which leakage from blood vessels in the back of the eye also causes photoreceptors to degenerate. Vision loss occurs in these later disease stages.
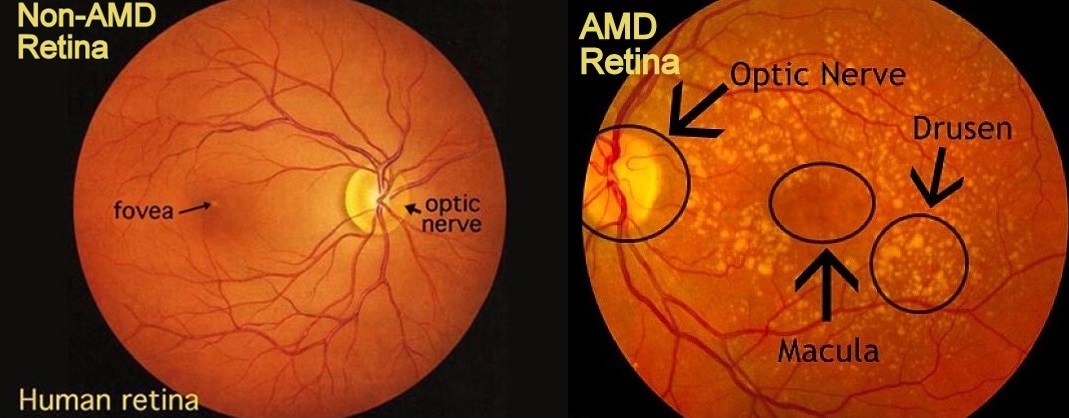
Figure 1. Images of Normal and Early AMD Retinas.
Lipid deposits in the membranes below the photoreceptors called drusen are an early sign of AMD. Usually they appear first in the periphery of the macula, such that central vision loss is not reported in early disease stages. Images from http://www.big-images.com and http://weluehealth.com, respectively.
The goal of the lysosome team is to find a way to counteract the atrophy that occurs in AMD. My summer project will involve building a model for studying dry AMD in RPE cells in culture. RPE cells provide nutrients to and process waste from the photoreceptors in the retina, which capture incoming light and begin processing it into the images we “see.” Currently, an RPE model for AMD does not exist, partially because researchers are not exactly sure what causes the cells to age and develop AMD but also because there is controversy in the scientific community regarding the characteristics of “aged” RPE cells. We will use three proteins identified as significant in AMD to assay how our RPE cells compare to aged AMD RPE cells by comparing our protein expression patterns to those in recent studies of the cells. The proteins we will use for these assays include a protein in the extracellular matrix of the RPE cells as well as transmembrane proteins implicated in faulty lipid transport. Researchers hypothesize that one of the causes of drusen formation and AMD is inefficient lipid transport in lysosomes, which may lead to the accumulation of photoreceptor waste in the underlying RPE and Bruch’s membrane. Additionally, we will use proteins that are markers of RPE cells to compare our model against other RPE cells we have in culture. Once the RPE model is developed, the lysosome team will use it to study the effects of a lysosomal enzyme, SENS20, on the aged RPE cells. This enzyme will hopefully restore function to the RPE cells.
Future Plans:
In the very near future, I am planning to apply to graduate school programs in genetic counseling. Hopefully my knowledge of the research process of developing treatments and cures for diseases with a genetic component will complement my understanding of the available and soon-to-be realized treatments for genetic diseases. Additionally, I hope my Summer Scholar project will build a new tool for extensive use in studying AMD and other types of macular degenerative diseases.


